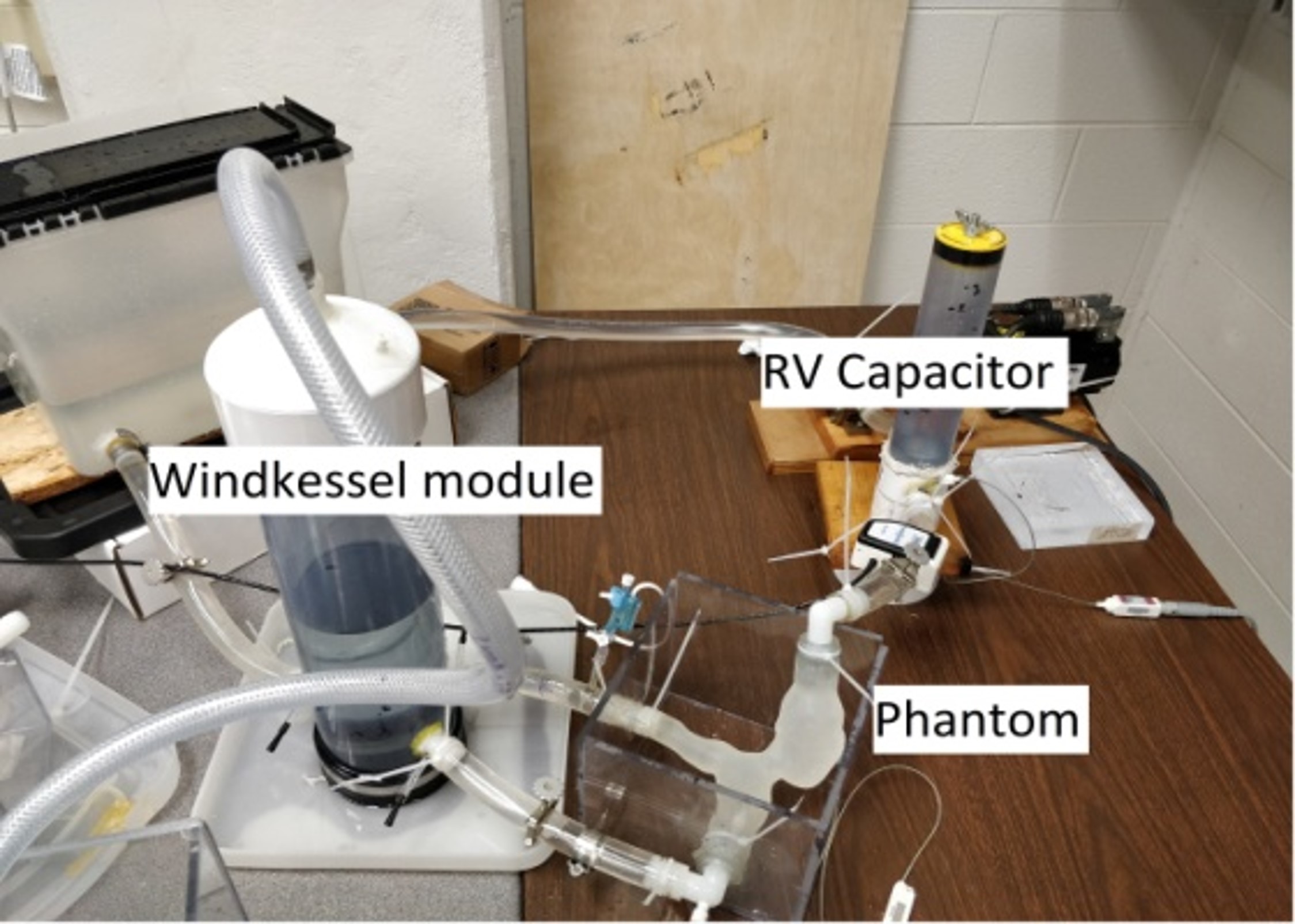
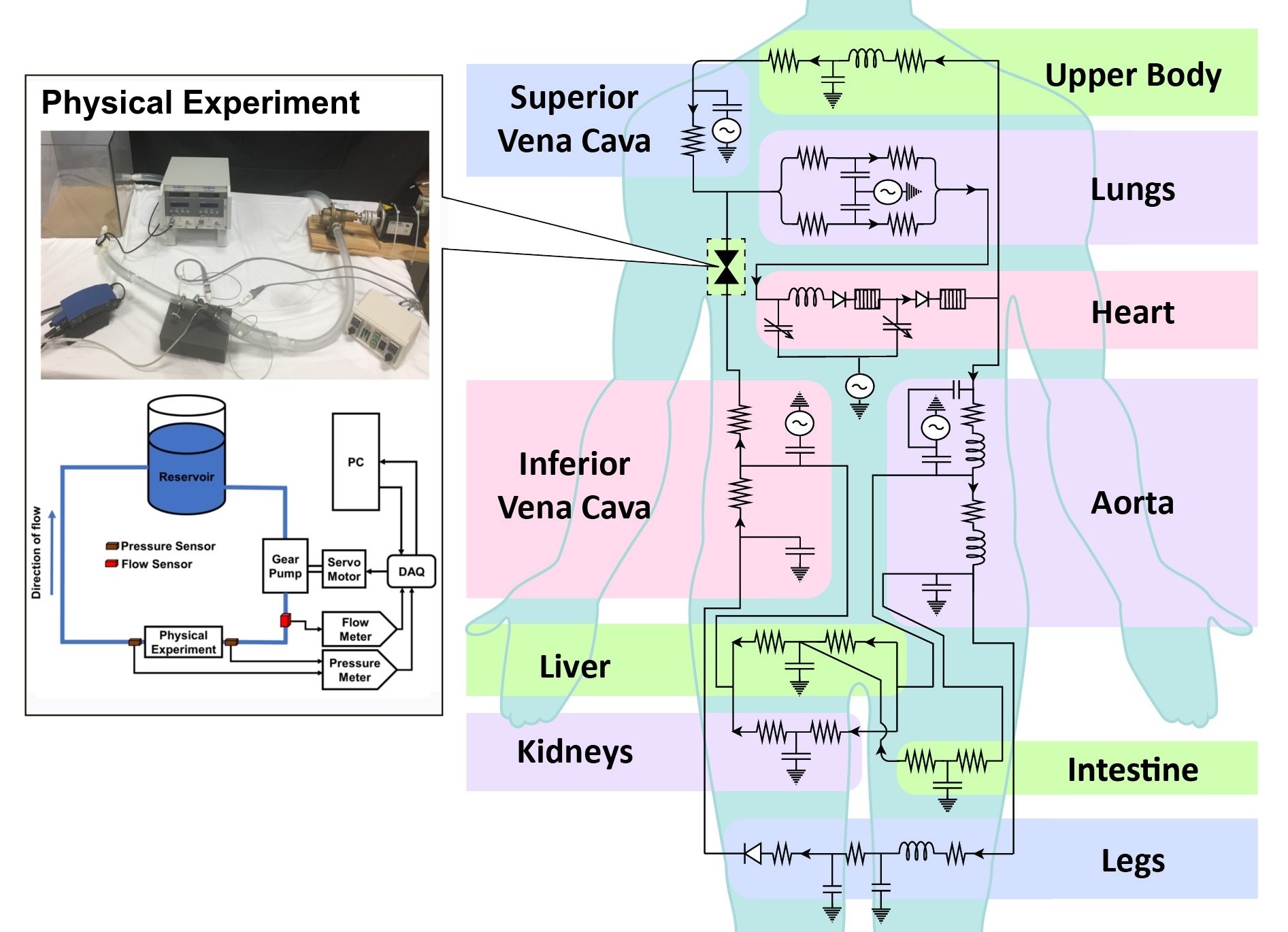
The behaviours of the cardiovascular system can be approximated via simplified circuit representations. Using this approach, we describe the effects of different cardiovascular system components such as contractility, vascular impedance, atrio-ventricular synchronization, auto-regulation, etc, and perform 0D simulations of pressure and flow to investigate disease conditions, medical procedures, and physiologic states.
Our lab conducts research to construct, improve, and validate these physiology models. This can involve applying regression/machine learning to analyze clinical or literature data, performing optimization to tune the relevant model parameters, and/or implementing mathematical models to describe a specific physical scenario, medical procedure, or device operation.
A few examples of clinical-focused research projects involving the application of physiology modeling: 1) We introduced dysfunctions such as limited heart rate and impaired respiration into a virtual patient to study the resulting exercise performance; 2) We simulated steady versus pulsatile ventricular assist in patient-specific models to investigate why steady flow devices tends to produce better outcomes in neonates; 3) We modeled a range of patient conditions to determine the optimal surgical configuration and device setting for a cavopulmonary blood pump.
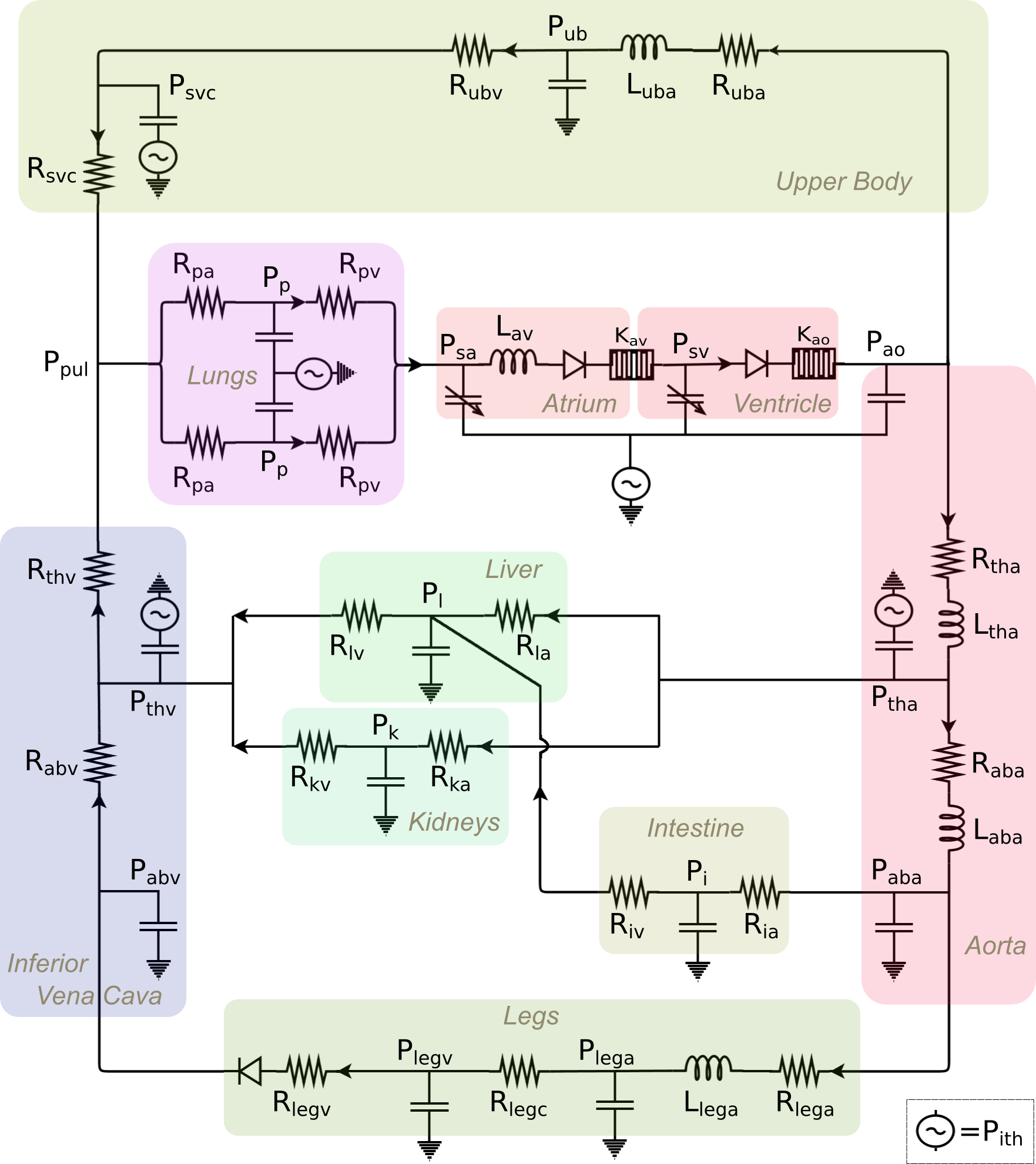 A lumped-parameter physiology model of the single-ventricle circulation
A lumped-parameter physiology model of the single-ventricle circulation